
Experimental Evolution: Identifying Pathways of Viral Resistance
By: Dasyre Sires Email: sire1910@vandals.uidaho.edu
Home Town: Moscow, Idaho High School: Kennewick
Major: Psychology
Department: Psychology/Communication Studies, Biological Sciences
College: College of Letters, Arts and Social Sciences, College of Science
Abstract
Phage therapy and antibiotics have been used in treating bacterial infections for nearly a century. However, bacterial mutations over time have allowed bacteria to advance in fitness and, in turn, resistance to these treatments. In this experiment we evolve phage-resistant bacteria to observe the mutations present on a genomic level. Pairing these observations with previous research regarding phage infection mechanisms will hopefully aid in the understanding and control of bacterial fitness.
From laboratory host Escherichia coli, we have isolated colonies that demonstrated resistance to each of 10 selected types of bacteriophage (Table 1.). We carried out plate evolution experiments with individual host colonies and each of 10 phage isolated from each of the bacteriophage strains. The genome of a resistant host from each experiment was sequenced in its entirety and analyzed for a total of 100 genomes. Sequencing artifacts where then identified in these colonies and compared to the ancestral strain to identify mutations. Similarities and differences in mutations are now being identified and analyzed to help us better understand factors of phage resistance.
Objective
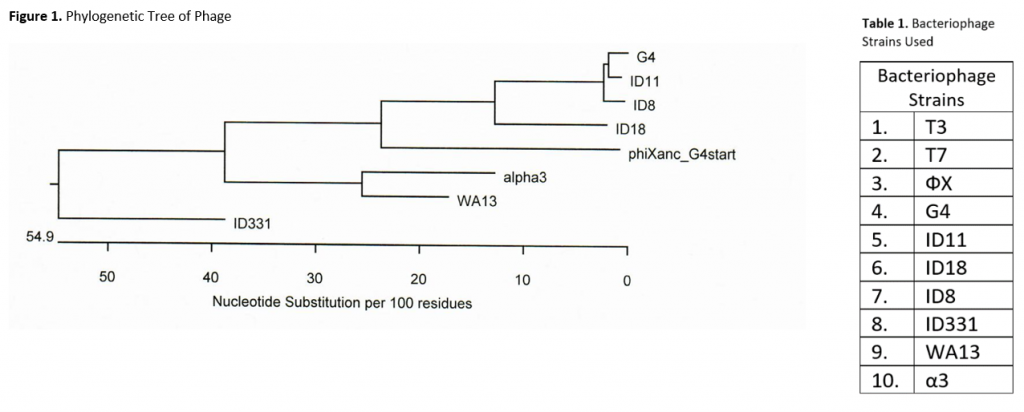
The objective of this project is to identify mutations in E. Coli C colonies resistant to one of ten different strains of bacteriophage. The bacteriophage strains we are using are listed in Table 1 and include eight ssDNA microviruses and two dsDNA phage, T7 and T3. This allows us to observe differences in mutations based on the phages’ infection mechanisms. Based on background research, I believe we will find more similar mutations between the host bacteria resistant to phage with more similar infection mechanisms. Additionally, I suspect that there will be a correlation between proximity of phage in the phylogenetic tree and similarities in mutations of the respective resistant E.-coli (See Figure 1). The tree below was made from a gene H (pilot protein) alignment and does not include T7 or T3.
I also would predict the infection mechanism of the phage type will partly determine which mutations are effective. Receptor mutations on the E. coli cell surface or enhancements in defense systems such as the CRISPR system may be more present against one type of mechanism.
Procedure
The procedure for this project has been broken down into two separate parts. The first part has just recently been completed and is outlined in the figure below. During this first part, we isolated colonies resistant to different phage types. After verifying resistance, these genomes where sent out for sequencing.
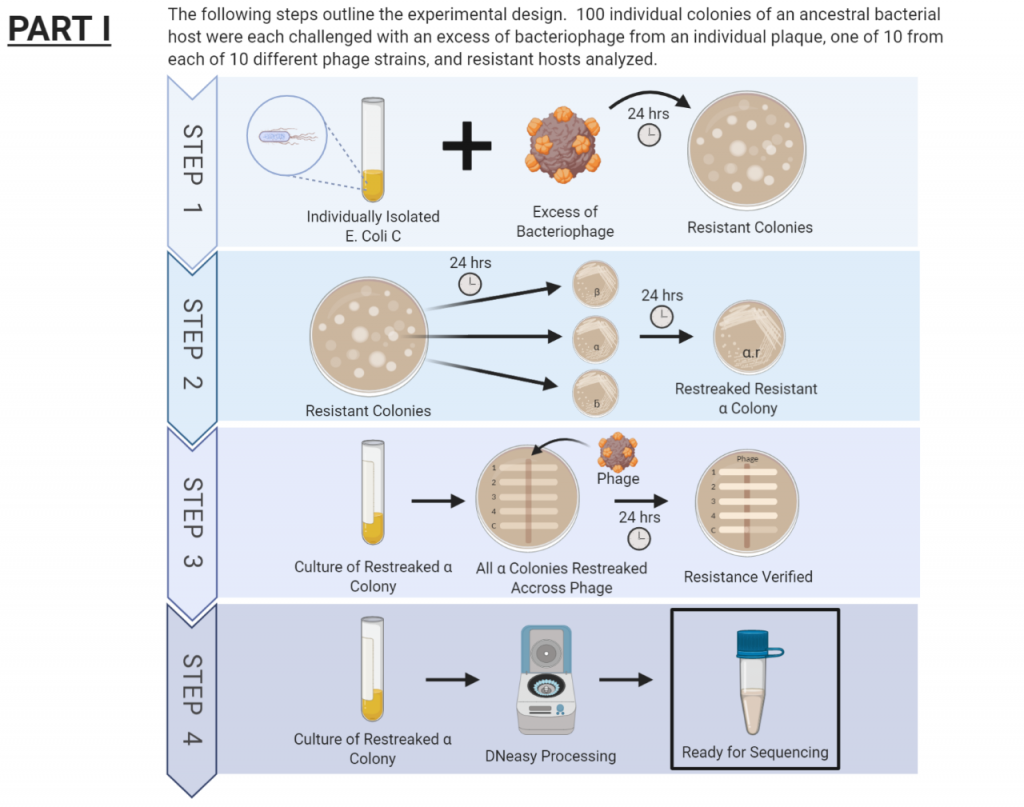
The second part of this project has involved working through the genome sequences of each resistant colony and comparing them to the ancestral genome. The discrepancies were flagged for further investigation to identify mutations. We are now in the process of identifying the various types of mutations and the effect of the mutation on the colonies. Understanding this will allow us to better infer how these mutations may be contributing to increased resistance.
Relevence
Ultimately, I hope that the results and analysis of this experiment will aid in understanding the various pathways of viral resistance or aid others in related research.
As will be demonstrated in this study, mutations that increase bacteria’s resistance to phage are not uncommon and may even be increasing. This presents concerns in regard to treating life threatening bacterial infections with phage therapy. This avenue of treatment utilizes phage’s ability to kill bacteria cells as a method of combating infection. This treatment strategy is gaining traction and becoming a very plausible alternative to antibiotics. However, if the bacterial cells are growing in resistance, then this approach will become increasingly ineffective.
Though antibiotics are another form of treatment, they often have more side effects than phage therapy. Unfortunately, antibiotic resistant mutations are also continuously evolving. This research may identify common pathways of resistance and could offer insight on ways we can reduce bacterial fitness from a general stand point. This would not just increase the effectivity of phage therapy, but antibiotic treatment as well. The results of this experiment will hopefully provide further understanding on how to combat bacterial resistance to bacteriophage and, in turn, help increase the effectivity of phage therapy. It is also possible that the experimental approach of this study and its findings may offer some insight for those looking to explore and study antibiotic resistance.

