Phage Engineering to Understand Virus Host Range
By: Zoe Wilson Email: wils6251@vandals.uidaho.edu
Home Town: Prosser, Washington High School: Prosser High School, 2015
Major: Microbiology
Department: Biological Sciences
College: College of Science
Viruses are the most common entities on Earth, outnumbering bacteria by around an order of magnitude, and animals/plants by another order of magnitude. Viruses are also the least well understood group in terms of diversity, function, and evolution. The goal of my project is to explore how viruses can exploit new hosts. Extrapolating what we learn in the lab with a simple, well-studied virus will help scientists predict the outcome of virus evolution in disease outbreaks and other naturally evolving virus populations.
ΦX174 is an icosahedral bacteriophage that is often used as a model virus due to its small genome and ease of cultivation in laboratory conditions. Although its natural host bacterium is unknown, it is thought to infect Escherichia coli and other mammal-associated bacteria with rough-O lipopolysaccharide (LPS) outer membranes, making ΦX174 ideal for studying the interaction of viruses with host LPS through experimental evolution. In this project, I am examining how mutations in ΦX174 affect LPS host specificity.
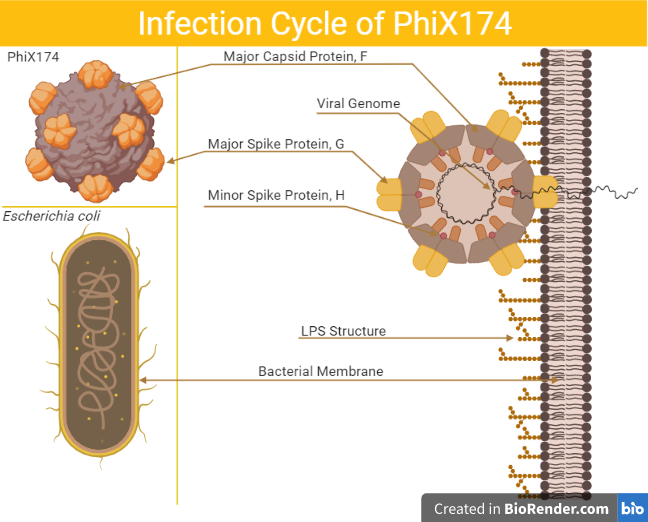
The F and G proteins of ΦX174 work together to recognize and bind to the host cell’s outer membrane. As virus entry is determined not only by the G protein, but also by the F protein, I am examining how mutations in both F and G proteins affect LPS host specificity.
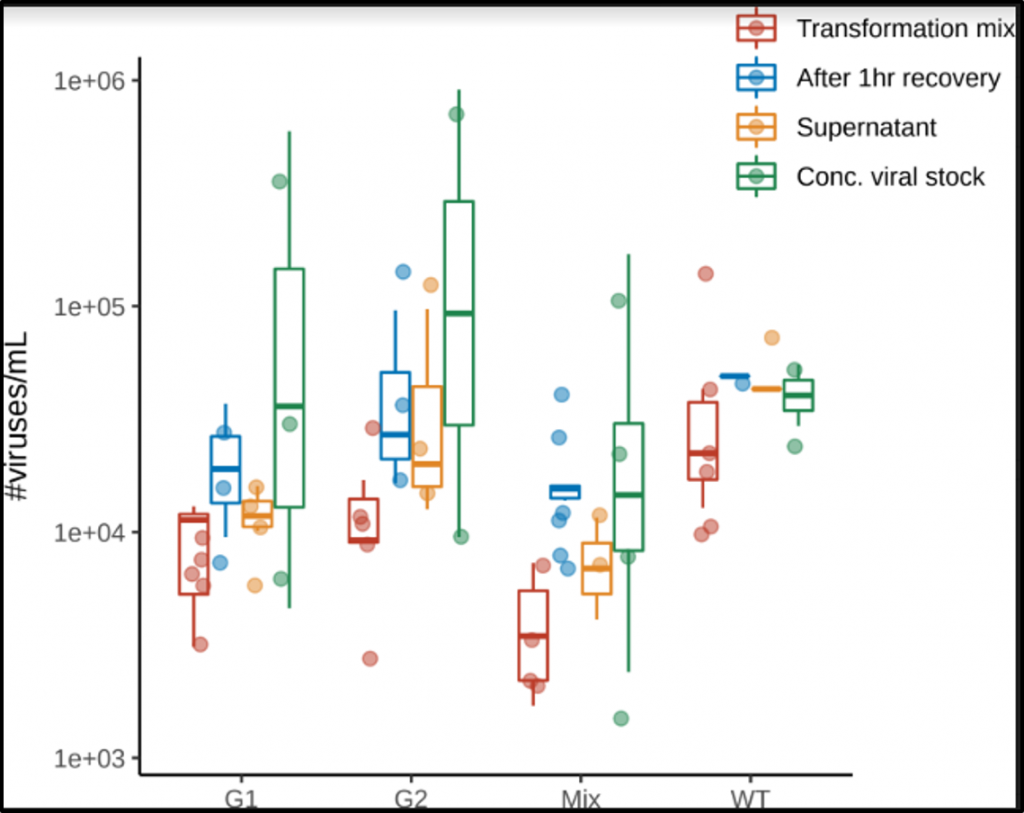
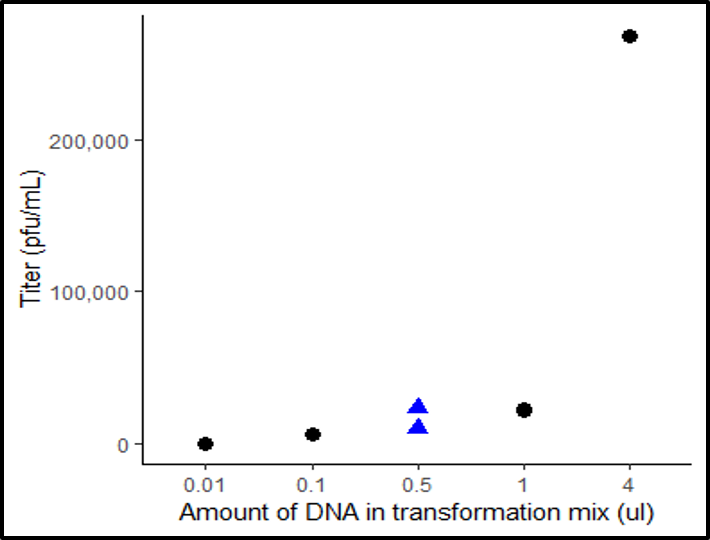
The lab has been investigating the effects of making mutations to the G protein in an attempt to build models that can predict the outcome of virus mutations occurring in natural populations. To do this, we make mutations to the G protein of ΦX174 and measure how well the mutant viruses grow. Over the past year, I have been helping to develop a high-throughput method of measuring viral fitness that competes many mutants against one another to test their fitness. The methods and initial results of this work have been published in a peerreviewed journal (ACS Synthetic Biology).
Images from: https://doi.org/10.1021/acssynbio.9b00411
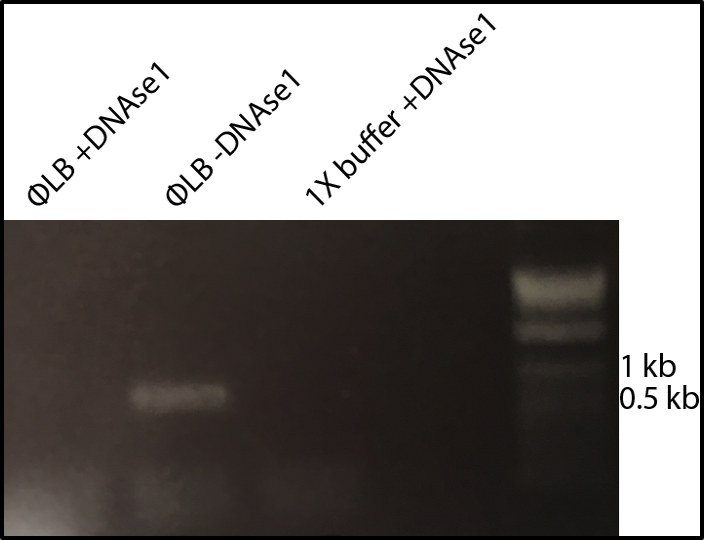
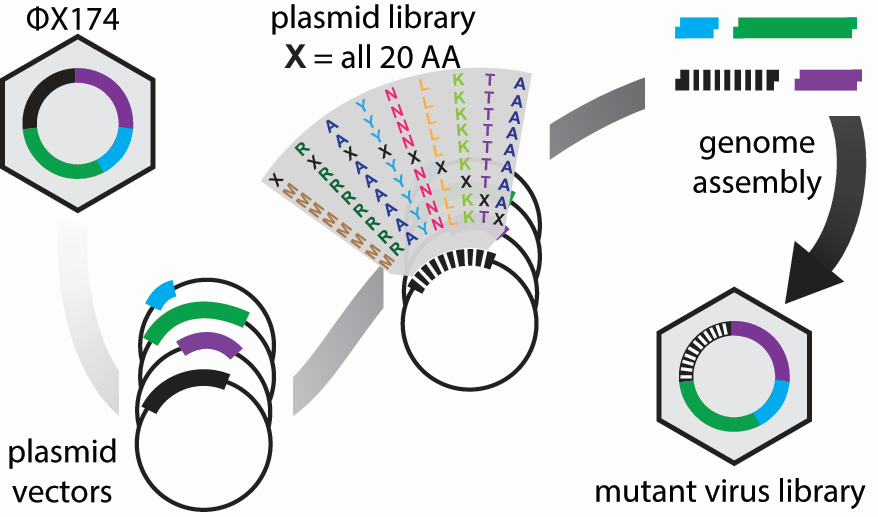
The high-throughput method involves making thousands of mutations to the virus, growing the mutated viruses in a flask together, and sequencing the DNA of the viruses at different time points to see which variants grow best. The lab has fragments of each portion of the phage’s circular DNA. These fragments are ligated together, along with a fragment containing mutations, to create the mutated phage.
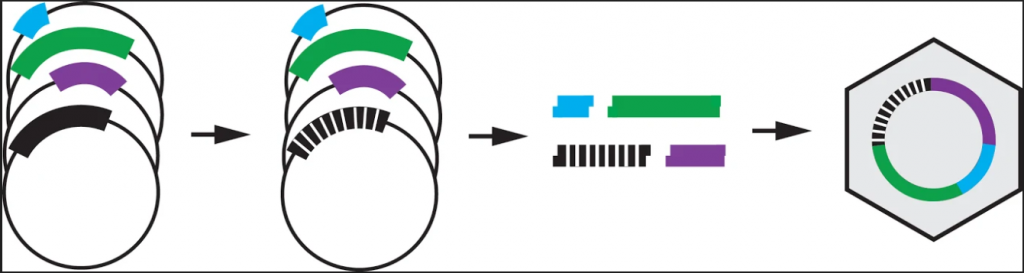
Image provided by JT VanLeuven
As a test for the high-throughput method, I have sequenced viruses with mutated gene G. I am currently comparing this data to traditional fitness data already collected for certain G protein mutants. This comparison is important for gaining confidence in the high-throughput method. Below is some data generated from sequencing data of the G protein.
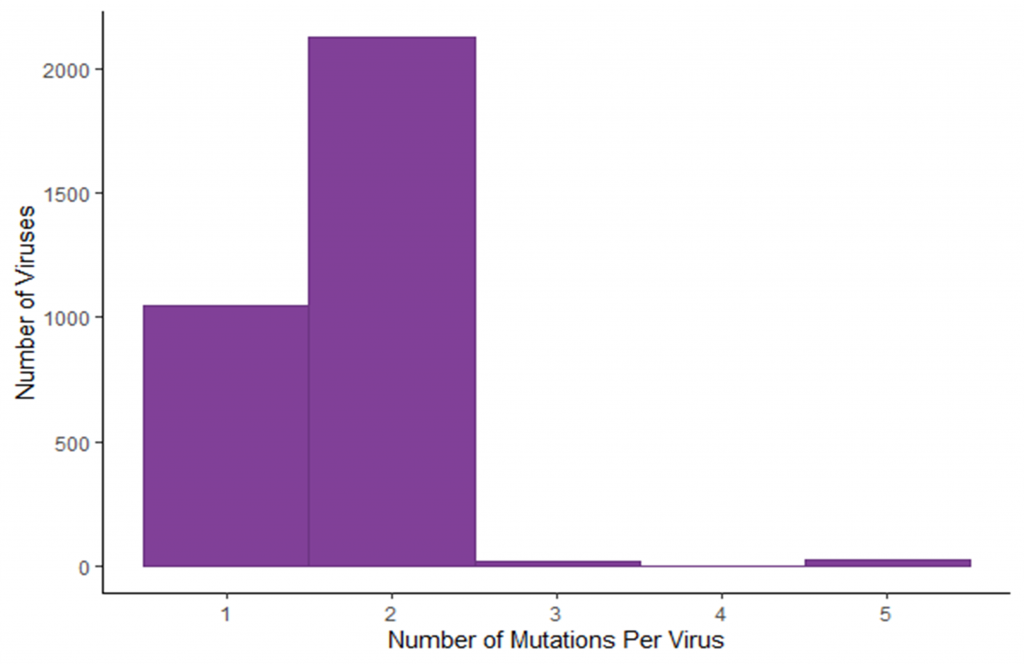
Image 2: Number of mutations identified in each virus. The majority of sequenced viruses had two mutations, as expected.
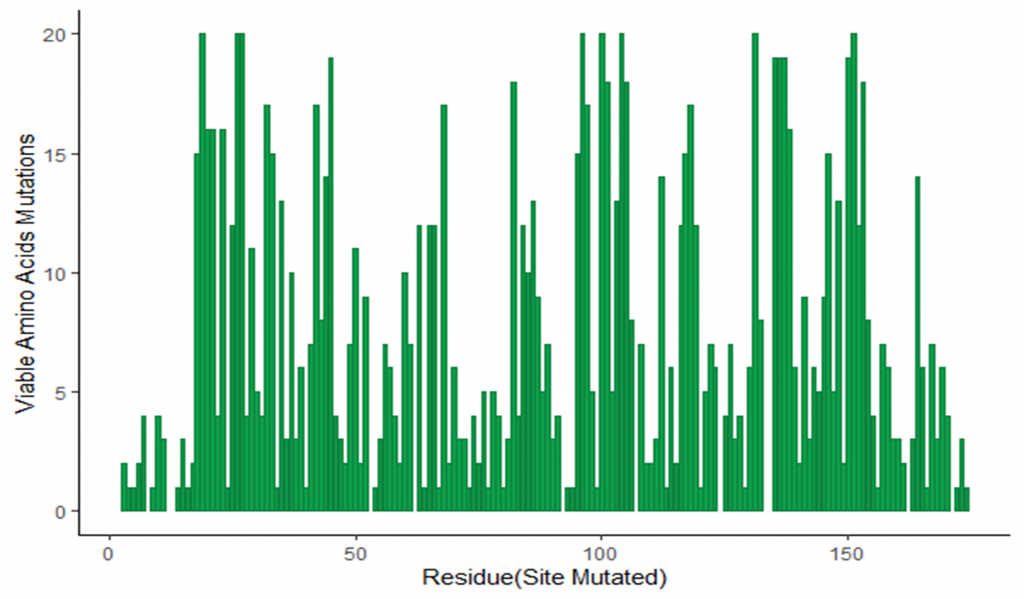
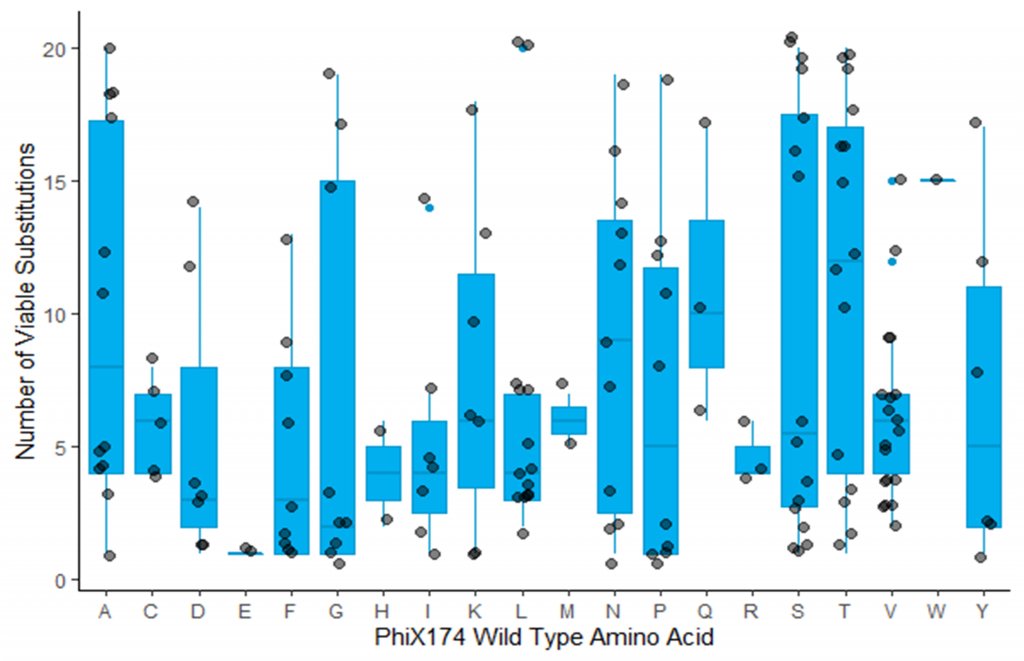
Image 3: Number of viable amino acid substitutions observed in G mutant library for each amino acid in the wild type virus.
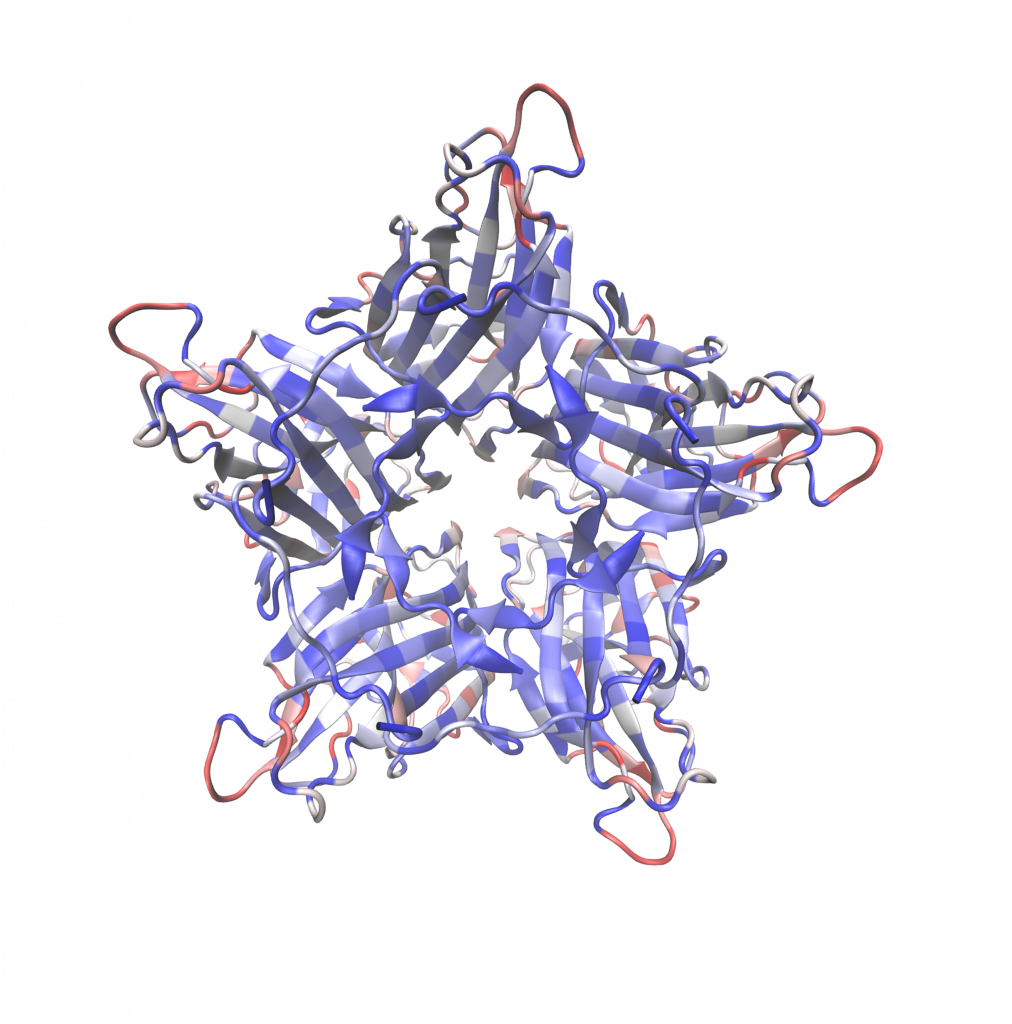
Image 4: 3D heat map showing frequency of observed viable mutations in G mutant
library at specific locations. The heat map is on a scale of 0 to 20. Red being all 20
amino acids observed, while blue represents no amino acid substitutions observed.
The structure of bacterial LPS can be diverse. Even within bacterial species like E. coli, strain-level differences in LPS structure may influence the infection/entry stage of the viral life cycle. The molecular interaction between LPS and virus proteins is thought to drive the very limited host range observed for most bacteriophage. However, previous studies have documented the evolutionary adaptation of ΦX174 to different hosts. Host switching often requires only a few mutations. For example, wild type ΦX174 acquires only a couple of mutations in gene F while adapting to Salmonella enterica. These mutations make the virus grow poorer on E. coli C. These types of studies, where a virus must evolve to grow on an alternative host give a limited view of the mutations conferring host specificity because not all mutations are equally accessible by evolution, especially when in competition with other mutations. Using the method that I helped develop, we can test the ability of every single possible mutation to grow on alternative hosts because we make the mutations instead of waiting for them to happen. The purpose of this project is to identify mutations in F that affect viral interactions with host LPS and to test for specific differences between LPS types by growing ΦX174 on several permissive and non-permissive strains of E. coli and S. enterica.
Products Produced:
| Type: | Title: | Date Published/Presented: | DOI: |
|---|---|---|---|
| Poster | Developing and Testing a High-throughput Protocol to Measure Viral Fitness | October 31, 2019 | |
| Publication | Saturation Mutagenesis Genome Engineering of Infective ΦX174 Bacteriophage via Unamplified Oligo Pools and Golden Gate Assembly | December 11, 2019 | https://doi.org/10.1021/acssynbio.9b00411 |
Additional Project Information:
Year in College Project Started: Junior
Faculty Advisor: JT Van Leuven
Faculty Advisor Email: jvanleuven@uidaho.edu
Funding Source: Hill Fellowship, DBS Undergraduate Research Grant: Karen Pohl Award
External Link to Project Information:
Project Location: Moscow, Idaho