
Segmentation analyses to identify and quantify microglia in 3D image stacks
By: Dan Blanchette Email: blan5568@vandals.uidaho.edu
Home Town: Coeur d'Alene, Idaho High School: Coeur d'Alene High School
Major: Computer Science
Department: Computer Science
College: College of Engineering
Abstract:
Using time-lapse confocal microscopy to record and observe microglia
behavior in living zebrafish embryos, the Mitchell lab investigates the
molecular basis of dynamic migration and phagocytic behavior of microglia
in the central nervous system. Microglial cells regulate brain development,
maintain neuronal networks, and repair neural injuries. Timelapse imaging
provides crucial insight into the behavior of these cells. Other methods, such
as manual cell counting, are subject to human bias and are tedious,
repetitive, and time-consuming. The Long computer science lab utilizes
Python and open-source modules to develop an automated pipeline as a
programmatic solution. These methods aim to expedite and optimize the
lab’s manual processes. This research project seeks to segment microglia
when applied to 3D time series image stacks. Computer segmentation of
microglia imaging relies on pixel values to generate bitmasks. Microglia
segmentation is challenging because the cell’s irregular shape and size can
be obscured, resulting in omitted pixel intensity values. Two programs were
developed to test the viability of Otsu, multi-Otsu, and Yen automatic
thresholding[2]. Both methods separate the raw image into two layers: the
foreground and background. Numerical data is generated via histograms to
predict optimal threshold values for each algorithm. Prototype
implementations of these programs demonstrate viability for microglia
counting. However, misclassification of microglia occurs when the predicted
pixel threshold is outside the histogram’s range. Further refinement of these
programs is crucial as they will be foundational for future object
classification methods and microglia tracking, optimizing the lab’s data
processing capabilities and time.
Background:
o Microglial cells regulate brain development, maintain neuronal
networks, and repair neural injuries.
o Bit masking is the method of isolating specific pixel color value
combinations (objects of interest) from the rest of a digital image.

o Previous versions of this program were applied to 2D images with a set
threshold value. This iteration aims to adapt dynamic
thresholding methods to multiple 3D image stacks and channels.
Materials
Software Packages
o Anaconda
o Jupyter Lab
o Spyder IDE
o Visual Studio Code
Python Modules/Dependencies
o Matplotlib
o Numpy
o OpenCV
o Sci-Kit Image
Program Version Control
o Google Drive
o Google Colab
o Github
Programming Languages
o Python 3.11.4
o BASH (Scripting Language)
Program Technical Specifications:
Hard disk space (programs and Spyder IDE): 659.1 MB
Operating System: PopOS! (Linux/Ubuntu 22.04 distribution)
Memory: 8GB, DDR4 SDRAM (recommended)
Compatible GPU: NVIDIA GeForce RTX 2060 6GB RAM
Methods
The figures and images for this section have been omitted from public release as requested by my faculty advisor as the image data has not been published.
Results
The figures and images for this section have been omitted from public release as requested by my faculty advisor as the image data has not been published.
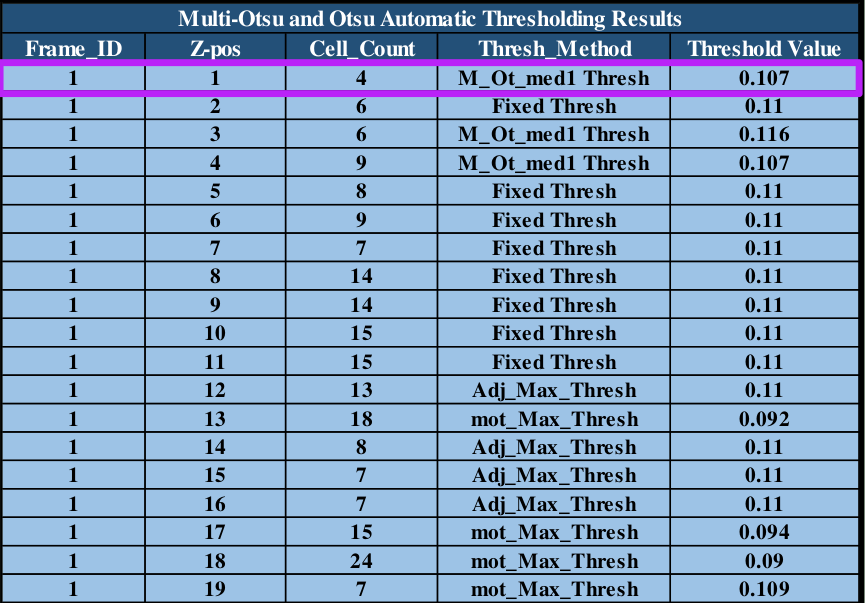
Table 1: program cell counts and threshold values for dynamic segmentation application.
Discussion and Conclusions
- The programs provide comparisons between image data sets.
- Further research and experimentation is required to improve results.
- The Yen algorithm performs well on images with a clear distinction
between pixel intensities for object segmentation. - The multi-Otsu approach allows for median pixel values to be
included as part of its segmentation. - The programs reduce time requirements to quantify microglia cells
which is one metric the lab uses to analyze microglia behavior. - Overall, program counts that are off by manual margins of error are
not detrimental to the laboratory’s data-gathering processes.
Future Work
- Research and development is required to reduce instances of cell
object misclassification. - Future object classification and tracking methods will be applied via
machine learning or A.I. algorithms. - Adding functionality to determine if cell objects of the same pixel
intensity and cartesian position should be counted or if it is part of a
larger 3D object. - Many experimental open-source image-tracking libraries(like
Pyclesperanto) are still under development but with updated
technologies may be applied to enhance microglia cell tracking while
maintaining open-source licensing for lab imaging.
Acknowledgments
- The Mitchell lab for their assistance with the biological science and terminology related to this
project. - Jordan Reed for her insights on testing multiple thresholding methods to enhance program
performance. - Professors Kirsten Blanchette, Rhena Cooper, Susanne Bromley, David Foster, and the North Idaho
College INBRE cohort for your feedback and support this summer. - Dr. Nate Schiele and the University of Idaho cohort for providing professional development
opportunities and project review. - My friends and family for their support and listening to me talk about my research over the
summer. - This project was made possible by SURF/INBRE
The project described was supported by an Institutional Development Award
(IDeA) from the National Institute of General Medical Sciences of the National
Institutes of Health under Grant #P20GM103408
References
[1] The Data Carpentry Family of Sites. Image Processing with Python, 2017,
https://datacarpentry.org/image-processing/01-introduction.html.
Accessed 23 June. 2023.
[2] Sreenivas, Bhattiprolu . “192 – Working with 3D and multi-dimensional
images in Python.” YouTube, uploaded by DigitalSreeni, 14 Jan 2021,
https://www.youtube.com/watch?v=kt8SZrSkGGw.
Accessed 23 June. 2023.

