Tailocin treatment of American Foulbrood Disease in honeybees
By: Katherine Ryssel Email: ryss8514@vandals.uidaho.edu
Home Town: Coeur d'Alene, Idaho High School: Coeur d'Alene High School
Major: Microbiology
Department: Biological Sciences
College: College of Agriculture and Life Sciences, College of Science
Tailocins are antibacterial proteins produced by some bacteria to kill closely related bacterial strains. In nature, this benefits the microbe by offering a way for them to control the bacterial composition of their environments. However, in a lab setting, researchers can use these tailocins to treat against unwanted bacteria. The purpose of this research project was to test whether a tailocin treatment is a viable option in protecting honeybees against the deadly American Foulbrood disease, which is caused by the bacterial species Paenibacillus larvae. To test this, the research involved three major steps which were intended to be completed across a 10-week period. The first three weeks were spent on collection and purification of hypothetical tailocins from strains of P. Larvae. The following six weeks were spent on in vitro analysis of the hypothetical tailocins against strains of P. Larvae. The final week was spent acquiring TEM images of samples to determine if any tailocin-like structures are present in the supernatant. The original timeline of this 10-week project involved the cell-free cloning of identified tailocin proteins followed by in vivo treatment of infected bees, but initial steps ended up taking much longer than expected. Further research will be done under alternative funding.
Honeybees are vital to the environment and the global food supply. They are responsible for pollinating many crops, providing a large portion of the pollination necessary to produce the fruits and veggies humans consume every day. However, honeybee populations have been declining due to several factors, including disease, the use of pesticides, climate change, and environmental destruction (Halvorson et al. 2021). One of the most significant threats to honeybees is the presence of pathogenic bacteria, which cause diseases that lead to the death of whole colonies (Forgesen et al. 2018). One example of this is American Foulbrood disease, caused by the bacterial pathogen Paenibacillus larva. Current treatment options for honeybees against P. larvae infection are limited and often ineffective. Antibiotics are currently allowed for use in the United States under the guidance of a veterinarian but are not approved in the EU. Moreover, antibiotic resistance is spreading in strains of P. larvae. Standard treatment in the EU is the complete destruction of infected hives by burning. The use of bacteriophages with specific targeting against P. larvae shows some promise (Jończyk-Matysiak et al. 2020; Tsourkas, 2020) but is not yet available. In addition, a recently approved vaccine against P. larvae provides a 30% reduction in mortality, but it is unclear if beekeepers will adopt this preventative measure. The purpose of this research was to explore the potential of utilizing tailocins as a treatment against these pathogens.
Bacteria have developed many ways to combat rival bacteria within their host or ecosystem. For example, some secondary metabolites in a bacteria can produce antibiotic activity which the cell itself has become resistant to but will damage or kill other nearby bacteria. Bacteriotoxic peptides and proteins, often called bacteriocins, tend to target bacteria that are very closely related to the producing bacteria. Tailocins are like these bacteriotoxic proteins in that they are sometimes able to target closely related bacterial strains with high specificity.
Some tailocins mediate interaction with a bacterium and the eukaryotic cells of its host, but the ones focused on in this study are those which are produced by bacteria to kill off similar, competing strains of that bacteria. This way, if tailocins can be isolated from Foulbrood-causing bacterial strains in bees from one colony, that tailocin stock may be used to cure an infection of Foulbrood in another colony.
Tailocins are a type of antibacterial peptide produced by bacteria which assemble themselves in similar fashion to phage tails without phage heads. It is considered likely that the gene clusters which code for tailocin production originated in bacteriophage. This is because the well-understood genes of F- and R-type tailocins appear to be highly similar in genetic makeup to the tail-assembly genes in phages of the Siphoviridae and Myoviridae families, only lacking the genes for phage head assembly and DNA Packaging (Maarten, G.K., Ghequire, René, De Mot). These similarities, along with others seen in various tailocin-producing bacterial families, indicate that the tailocin genes clusters did not evolve independently. However, tailocin clusters are not actually considered prophages. To be considered a prophage, the sequence of DNA must have sequence matches to a core, tail, and integrase, and also have a sequence denoting lytic ability. If the DNA section has all of these except for an integrase, then it is considered a bacteriocin rather than a prophage. This is the case for tailocins. (Derrick E. Fouts). It is more likely that tailocin gene clusters evolved from prophage in bacteria which experienced natural selection against self-targeting tail fibers in favor of tail fibers which attach to and kill competing bacteria (Carim, S., Azadeh, A.L., Kazakov, A.E. et al).
F(flexible)- and R(rigid)-type pyocins are the two best understood tailocin structures. They were originally distinguished in studies of Pseudomonas aeruginosa and are commonly used as models for any phage-like bacteriocins discovered. R-type tailocins consist of a core and sheath and are contractile (meaning the sheath will contract upon attachment to the target, inserting the core into the cell membrane). F-type tailocins have no sheath to keep them rigid, so they are instead highly flexible (Daniel Walker, Hannah M. Behrens, et al). Both structures are capable of binding to and lethally disrupting the cell membrane of the target cell by injecting a core or needle into the membrane and negating the bacterial membrane potential, which will cause depolarization or hyperpolarization of the membrane, resulting in cell death.
The specificity of tailocins is largely determined by the surface cell receptors on the target bacteria and how they match up with the receptor-binding proteins on the tailocin’s tail fibers. Most R-type tailocins have only one gene encoding for a tail fiber, so each tailocin type can usually only interact with one receptor type (Sascha Patz, Yvonne Becker, Katja R. Richert-Pöggeler, et al). F-type tailocins, and many other tailocins, often have more than one of these genes. To find a tailocin that could effectively be used as a treatment for Foulbrood outbreaks, we wanted to find one or more which have a broad target range for strains of P. Larvae but is not lethal towards other bacteria which are beneficial to a honeybee’s microbiome. A very low target-range tailocin would likely only be useful in a lab setting and would not be effective against a real outbreak, but a very high target-range tailocin would likely heavily disrupt the microbiomes of the honeybees and cause other negative effects on the hive. Effective uses of tailocins have been shown in many past studies, However, the tailocins of P. larvae and M. plutonis bacteria have not yet been studied, thus the potential for their use as a treatment against American Foulbrood in honeybees is unknown.
Induction of tailocins from P. larvae strains and in vitro assessment of host range. Overnight cultures of seven P. larvae bacterial strains were prepared by inoculating 3mL of media with a single colony, then incubated with shaking at 37°C for 16±2 hours. A 1:100 dilution was made from overnight cultures into 10mL of media, and allowed to incubate, shaking at 37°C to mid exponential phase. Test cultures were then exposed to ultra-violet light (254nm) for two minutes to induce the production of tailocin particles. After a few tests using UV light, it was determined that this stressor was not working well. Instead, cultures were induced with 1ug/mL of Mitomycin C. A negative control group was not exposed to a stressor. The cultures were then incubated overnight at 30°C to allow for the potential production of tailocin particles. Cultures were subjected to low-speed centrifugation at 5000 x g for 5 minutes to remove bacterial debris, and the supernatant was filter sterilized (0.22mm). Negative controls for each strain were processed in the same manner.Filter sterilized supernatant from induced and the non-induced control group of P. larvae strains were serially diluted and spotted onto lawns of recipient P. larvae bacterialstrains. Plates were incubated overnight at 37°C and observed for inhibitory activity the following day.
TEM imaging of samples. Two mL of each active sample were placed in centrifuge tubes and spun at 20,000 rcf for 2 hours. The supernatant was drawn off and kept for future tests, and the pellet was resuspended in 1 mL of pure water. This new mix was once again spun at 20,000 rcf for 2 hours. The supernatant was then discarded, and the pellet was resuspended in 100uL of pure water. Ten uL of each sample were spotted onto bacterial lawns to check if spun down samples would still show clearing activity. Remaining samples were then sent to Washington State University’s Franceschi Microscopy and Imaging Center for transition electron microscope imaging. To prepare for imaging, 4uL of a sample was placed onto a copper grid and treated with 4uL of uranyl acetate followed by washing with 4uL of water three times to create a negative staining effect. Grids were allowed to dry before they were placed into the electron microscope for imaging. Only three samples (368W, 25747, and 3670) were imaged. Two grids of each sample were made in case any grid had failed stain.
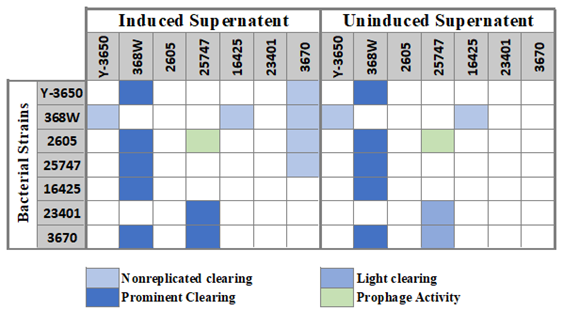
Induction of tailocins from P. larvae strains and in vitro assessment of host range. Of the seven P. Larvae strains which were tested, five showed tailocin-associated activity. Of those five, two had consistent and replicable results. Possible tailocin activity is considered when a sample creates a zone of inhibition on a bacterial lawn. This clearing can be easily distinguished from the plaques created by a prophage. When diluted, a prophage sample would create multiple smaller plaques on the lawn. A bacteriocin (such as a tailocin) would not create individual plaques, and instead, diluted samples show a faded zone of inhibition resulting from a less efficient killing of bacteri a in the area. This kind of activity was seen from strain 368W against 5 of the 7 strains. One of the strains it did not show activity against was itself, which was expected as producing strains are resistant to their products. Strain 25747 showed inhibitory activity against two strains, including one of the strains that 368W was ineffective against. 25747 also showed prophage activity against strain 2605. Additionally, other strains showed activity under certain conditions. Strains Y-3650 and 16425 both showed very light activity against 368W when lawns had very low bacterial concentrations. Strain 3670 showed activity against 4 or 5 other strains in two tests, but such activity has not yet been consistently replicable.
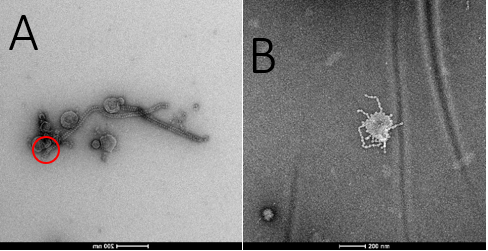
TEM imaging and analysis of samples. Three samples were taken in for TEM imaging. Only two of these samples were clear enough to take pictures. Sample 368W (left) had structures which somewhat resembled F-type tailocins, but it may also be a virus. No tailocin-like structures were able to be found in sample 3670 (right), although some interesting structures were seen which could not be identified. Sample 25747 had not been properly purified, and therefore was impossible to read on the TEM. Although samples 368W and 3670 were clear enough to take images, they were still very thick and had many large clumps of molecules which could have been hiding important structures. In the future, a better method of isolation must be found prior to imaging. It may be important to find an imaging facility which has the capability to run a glow discharge on the copper grids which samples are placed on for imaging. Glow discharge may prevent the clumping of certain charged molecules and therefore allow clearer images to be taken.
Activity from tailocin proteins produced by 368W show the most promise for treatment of infected bees. Although 25747 showed more prominent clearing than 368W, 25747 had a far more limited host range, showing activity against only 2 of the 7 tested strains while 368W showed clearing against 5 of the 7 strains. In the two trials where activity from 3670 was observed, 3670 showed both a high host range (4 of 7 strains) and very strong killing activity, which would make it a strong contender if activity was more easily replicable. In the first trial in which activity from strain 3670 was observed, Mitomycin C was used for the first time (instead of UV light) to induce tailocin production. The second time in which activity was observed, a doubled concentration of Mitomycin C was used. This may mean that 3670 requires a high amount of stress to begin producing tailocins and that, upon opening a container of Mitomycin C for the first time, the drug began to decay. This would explain a higher concentration being needed to induce production again.
Because portions of this project took longer than expected, there are still many more steps to take in the future. Big picture goals include cloning tailocin proteins and running in vivo tests on infected bee larvae. Before research gets to that point, many other questions should be answered. Production of tailocins by strain 3670 should be repeated until activity becomes consistent. New preps for TEM imaging should be tested to get clearer images and hopefully locate the proteins responsible for killing activity in each sample. Additionally, there are some smaller questions which could be answered easily in the future such as the effects of pH, heat, or different forms of cell lysis (chloroform, sonication, etc) on the proteins responsible for killing activity in samples.

